In my previous post I created a Shiny app for exploring the text of Australian Prime Minister Scott Morrison’s speeches and interviews. While I was pleased with the app, one thing I didn’t like was having to run scraping and processing scripts manually to keep the data feeding the app up to date.
This post describes how I used GitLab CI/CD and and AWS S3 to automate my scripts and save their outputs so that I could use them in the ScoMoSearch app. Because this involved learning a few tools that were new to me, I’ll go through in detail the steps I took to get this working.
The original workflow and why it needed to change
The Shiny app was deployed using AWS EC2 and Shiny Server (following Charles Bordet’s excellent and comprehensive guide). Because the raw transcripts from the PM’s website require a lot of cleaning and processing, I opted to do these steps outside the Shiny app, save the R objects required, and then have the app load the saved objects. My initial workflow for updating the data feeding the Shiny was:
- Run scraping, cleaning, and processing scripts locally. The resulting objects required for Shiny are saved.
- Push to ScoMoSpeeches repo.
- Log into AWS EC2 instance running Shiny Server.
- Pull from ScoMoSpeeches repo. The deployed Shiny now has the up-to-date objects.
Though this wasn’t a complex workflow, it became tedious to perform regularly (particularly because the first step would take about 20 minutes on my laptop). After a week or so of doing this, it became clear that the process needed to be automated.
The approach I settled on required three major changes:
- Save R objects required for the Shiny in AWS S3 instead of locally.
- Have the Shiny read these R objects from S3. This means I don’t need to update anything in the EC2 instance for the Shiny to have access to current data.
- Run the processing scripts using GitLab CI/CD, which allows me to schedule them to run every day.
Storing and retrieving data in AWS S3
The first step towards making maintenance of the ScoMoSearch Shiny less painful was to remove its dependence on locally stored objects. What I wanted was to be able to update the data feeding the Shiny without updating the Shiny itself.
Enter AWS S3. This is a service that allows you to store files in the cloud. Files are stored in ‘buckets’, which are basically directories and can be made public or private. The AWS free tier allows you to store 5 GB of data. There are limits on the number of times you can access your bucket or add/update things, but this was well above what I needed.
To use AWS S3, you need to set up an AWS account and then generate credentials using the Identity and Access Management page. I followed the instructions in step one from this guide by Ben Gorman. Be aware that the only opportunity to download your credentials is immediately after generating a user, so be sure to get those and keep them somewhere safe. You will need both an AWS secret access key and an access key id. To access your bucket you’ll also need to know your region.
The aws.s3 package in R allows you to save and retrieve files from your S3 bucket. Importantly, your AWS credentials need to be provided to aws.s3 to connect to your bucket. However, you don’t want your credentials to be public so they can’t go in your script directly.
In the tutorial above they describe how doing this by setting environment variables. However, I couldn’t get this to work using Shiny Server, though it does work when running the Shiny locally.
Instead I stored my credentials in a .dcf file in the same folder as my Shiny and processing scripts. The format of a .dcf file is a set of key value pairs, which lends itself to storing variables. You can create a .dcf file in R using write.dcf() and read from it using read.dcf().
I added the .dcf to my .gitignore file to ensure it wasn’t accidentally made public when I pushed to my repo. Even if your repo is private, it’s not good practice to have passwords and other credentials in code.
Having set up S3, I then needed to change my cleaning and processing scripts to save the objects they made to S3 rather than to a local folder. I also needed to get the Shiny to read from S3 rather than a local folder.
Fortunately, saving an object (as an RDS) to S3 using aws.s3 is almost as easy as saving an RDS locally:
s3saveRDS(x = object_to_save,
bucket = "my_bucket",
object = "object_to_save.rds")You can then access the object using s3readRDS():
loaded_object <- s3readRDS("object_to_save.rds",
bucket = "my_bucker")(Optional step) Running scripts using Docker
Ultimately I wanted to run my scripts in a Docker container using GitLab CI/CD. However, having never used Docker or GitLab CI/CD before, I decided that getting my scripts to run in Docker on my computer was a reasonable intermediate step. This made it a lot easier for me to iron out any problems and get everything working before moving to GitLab CI/CD. Though it isn’t essential, I highly recommend this step if you’re still learning your way around Docker.
The official documentation provides a great overview of Docker and instructions for getting started. For using Docker with R, I closely followed Colin Fay’s post.
Docker allows you to work within a defined and isolated environment known as a container. Because the container is highly defined, it doesn’t matter if if the container runs on my computer or someone else’s (even if the operating system is different), as in both cases the container will be the same. This is excellent for ensuring reproducibility and is one of the main reasons Docker is used.
A container is a specific instance of an image. An image is basically a template for the container that sets up everything you need (for example, by installing packages and running scripts), so that when you start a new container all of that will happen automatically.
An image is built from a set of instructions called a Dockerfile. This is the Dockerfile I used for running my processing scripts, which I will go through line by line to explain the different elements:
FROM rocker/r-ver:4.0.3
RUN mkdir /home/analysis
RUN apt-get update \
&& apt-get install -y --no-install-recommends \
libxml2-dev
RUN R -e "install.packages(c('rvest', 'magrittr', 'data.table', 'aws.s3', 'stringr', 'zoo', 'sentimentr', 'textclean', 'quanteda'))"
COPY .aws /home/analysis/.aws
COPY 01_scraping.R /home/analysis/01_scraping.R
COPY 02_cleaning.R /home/analysis/02_cleaning.R
COPY 03_processing.R /home/analysis/03_processing.R
COPY 04_combine.R /home/analysis/04_combine.R
CMD cd /home/analysis \
&& R -e "source('04_combine.R')"My image will actually be built on top of another image. In the first line, I specify which image I want to use from Docker Hub (a collection of Docker images) using the keyword FROM:
FROM rocker/r-ver:4.0.3It’s very unlikely that you want to begin with a blank slate; in my case, I need to run R scripts so I chose an R-based image. These are available through Rocker (a collection of Docker images for R). For the sake of reproducibility, I’m using an image with a specific version of R. There are many difference R-based images available through Rocker, including many with R packages already installed.
In the next line, I create a new directory where I will move my R scripts before running them. The command is the same as you would use in the command line, but preceding by the RUN keyword:
RUN mkdir /home/analysisNote that this directory will be created within the container, so when I stop the container it and its contents will no longer exist. This is fine in my case since I’m saving the resulting objects in S3, but if you do need to transfer outputs to your computer Colin Fay’s post explains how to do this using volumes.
Next, I again use the run keyword to install the packages needed for my scripts:
RUN apt-get update \
&& apt-get install -y --no-install-recommends \
libxml2-dev
RUN R -e "install.packages(c('rvest', 'magrittr', 'data.table', 'aws.s3', 'stringr', 'zoo', 'sentimentr', 'textclean', 'quanteda'))"The first command installs libxml2 using the command line, which is required for the R package rvest. The second command installs the R packages required by the R scripts I will be running. The command R -e makes R execute a command from the command line. The backslashes simply allow me to add line breaks for readability.
Next, we copy the R scripts and the AWS credentials file to the previously created directory using the COPY keyword.
COPY .aws /home/analysis/.aws
COPY 01_scraping.R /home/analysis/01_scraping.R
COPY 02_cleaning.R /home/analysis/02_cleaning.R
COPY 03_processing.R /home/analysis/03_processing.R
COPY 04_combine.R /home/analysis/04_combine.RUp to this point, each part of the Dockerfile will be run only when the image is built and not each time the container is launched. However, some parts we will want to run every time. These commands are preceded by the CMD keyword:
CMD cd /home/analysis \
&& R -e "source('04_combine.R')"Here, we move to the directory containing the R scripts and then run them using R. The script 04_combine.R sources the other processing scripts, so it’s the only one to be called explicitly in the Dockerfile.
Now that the Dockerfile is set up, we build it using docker build -t analysis ., where analysis is the name of the image. This can take a while to build, depending on how much is to be installed. Then to start the container and run the scripts, use docker run analysis.
Though this step wasn’t strictly necessary to getting my scripts to run using GitLab CI/CD, it did help me understand better how Docker works and what exactly I need my GitLab pipeline to do. This made the next step a lot more straightforward.
Using GitLab CI/CD to automate scripts
To run my processing scripts automatically, I used to GitLab CI/CD to run them in a Docker container. You can read more about this in the GitLab documentation here.
To setup CI/CD, from your GitLab repo click the ‘Set up CI/CD’ button:

After clicking ‘Create new CI/CD pipeline’ on the next page, you will see a template for the .gitlab-ci.yml file that defines what will actually happen in your CI/CD pipeline.
The template is more complicated than the one I actually used, but illustrates the basic structure and how a pipeline can include several stages:
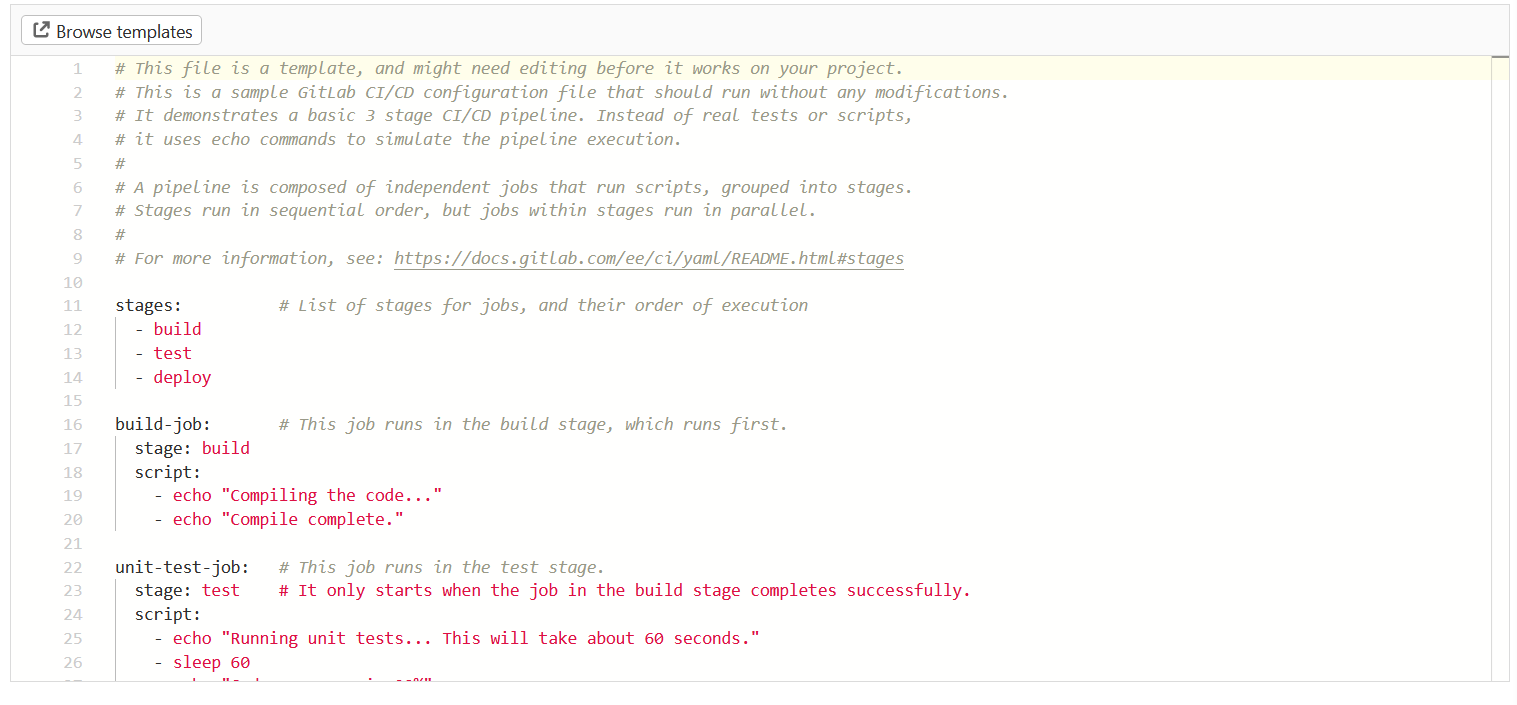
This is my .gitlab-ci.yml:
image: rocker/r-ver:4.0.3
execute:
script:
- apt-get update && apt-get install -y --no-install-recommends libxml2-dev
- R -e "install.packages(c('rvest', 'magrittr', 'data.table', 'aws.s3', 'stringr', 'zoo', 'sentimentr', 'textclean', 'quanteda'))"
- R -e "source('04_combine.R')"
This has several similarities to my Dockerfile above. The line image: docker:rocker/r-ver:4.0.3 tells Docker what image to use (similar to the FROM line in the Dockerfile).
The part under execute should look familiar from building and running our Docker image earlier. script is the name of the pipeline stage (in this case I only have one stage, so the word ‘pipeline’ is perhaps a little grand), and the following lines are terminal commands. The first two commands ensure that everything needed to run the scripts is installed, and the final one runs the processing scripts.
As when I was running them locally, the scripts require my AWS credentials to save generated R objects to S3. Instead of adding my credentials to GitLab and reading them from there (which would be a bad idea, particularly since this is a public repo), I added them as GitLab variables.
To do this, click ‘Settings’ in the sidebar and then ‘CI/CD’ in the expanded submenu. Under ‘Variables’, you can click ‘Add variable’ to add a key value pair. Be sure to click the checkbox ‘Protect variable’ if this is something that needs to be secret, like a password.
The final step is to schedule the pipeline to run (though ‘CI/CD’ and then ‘Schedules’ in the sidebar). Then simply add a short description and choose when you want the pipeline to run:
You can see the details of each time your pipeline has run though ‘CI/CD’ and then ‘Pipelines’ in the sidebar.
And with that, I never had to manually run one of my ScoMo transcript processing scripts again (in the few weeks since I’ve done this, at least). A very happy ending!
Resources
- I used the
aws.s3package to interact with S3 from R. - This guide by Ben Gorman on using S3 with R has a very clear walkthrough of how to set up S3 and how to use the
aws.s3package to save and load data from S3. - Colin Fay’s post on using R and Docker is great for getting started with Docker.
- The ScoMoSearch Shiny app now updates automatically once a day.
- All scripts I used to scrape, clean, and process the transcripts and the code for the Shiny itself are on GitLab.